Whole Genome Sequencing
With next-generation sequencing, de novo genome assembly for a new species, strain, or transgenic line is no longer a huge undertaking as it used to be. As of September 2020, more than 55,000 organisms have already been sequenced.
Most whole genome sequencing (WGS) projects are now focusing on genome re-sequencing. The goal here is to identify various types of sequence variants and their associations with diseases or phenotypes. Discovery of such variants, including single nucleotide variants (SNVs), insertions and deletions (indels), copy number variants (CNVs) and segmental rearrangements, is central to precision medicine, pharmacogenomics, and other branches of bioscience.
NUSeq deploys different WGS strategies to meet the needs of different projects depending on genome size and project objective. Detection of SNVs and short indels in small genomes is relatively straightforward. Identification of CNVs and segmental rearrangements in large genomes is more challenging. Recently we have adopted the Transposase Enzyme Linked Long-reads sequencing technology, or TELL-Seq, to provide scalable WGS for such challenging applications.
 |
|
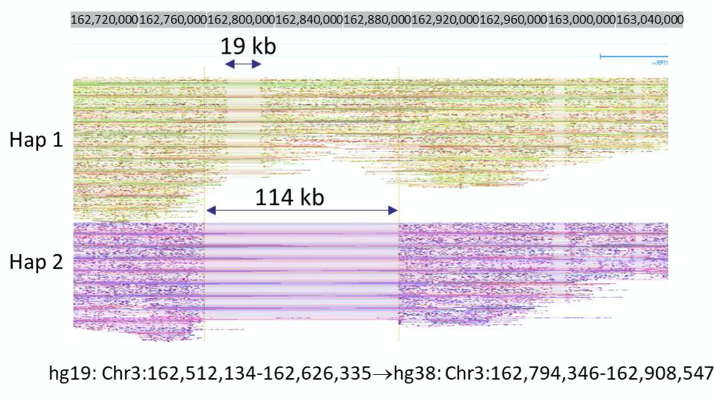
Detection of structural variation using TELL-seq. Phased reads generated from TELL-seq was used to uncover a 19-kb homozygous deletion within a 114-kb heterozygous deletion on Chr. 3. (Figure from Genome Res. 2020. 30: 898-909) |
Whole Genome Sequencing
Library Prep
A variety of WGS library construction approaches will be used, including Illumina DNA Prep, Illumina DNA PCR-Free Prep, Nextera XT, and TELL-Seq WGS Library Prep (for linked long reads generation). These different approaches are generalized as “DNA-Seq Library Prep” on our pricing structure.
Sequencing Mode
Selection of sequencing mode will be dependent on project specifics. NUSeq sequencers generate up to 300 bp paired end reads. With the use of linked long reads technology, structural variants detection or de novo genome assembly can be accomplished. For cost of the different sequencing modes, please check our pricing structure.
Service Request
Project consultation is provided free-of-charge. DNA-seq services can be requested through NUcore. Please finish and submit our project intake form when requesting service.
Sample Submission
The amount of genomic DNA needed varies with library prep approach:
- Illumina DNA: small genomes (e.g., microbial), 10-500 ng; large genomes (e.g., mammalian), 200-700 ng
- Illumina DNA PCR-Free: 30-500 ng
- Nextera XT: 5-10 ng DNA
- TELL-Seq WGS: 1-10 ng HMW (>20Kb) DNA
Please note that for DNA quantification, fluorometric-based methods, such as Qubit or PicoGreen, are preferred. Spectrophotometric-based methods, such as Nanodrop, may not be accurate. If there are excessive RNA in the DNA sample, it should be removed with a RNase treatment.
Bioinformatics
Data analysis is provided upon request. For variant analysis, standard DNA-seq bioinformatics service includes sequencing data QC, alignment and variant calling. Delivered results are variant call (VCF) files. De novo assembly is available. For large genomes, this may involve significant staff time charge.
Bioinformatics is billed on a per hour basis. Usually the time spent on a typical project is averaged at one hour per sample for the standard service.
Data Storage Policy
DNA-seq data is stored on the NUSeq server space for one year from the date of generation. It is recommended to transfer data files to the user's own space after they are generated.