Single-Cell Sequencing
NUSeq has offered single-cell sequencing since 2017. This state-of-the-art technology offers unprecedented opportunities to study cell-to-cell variation, identify/visualize different cell types/identities in a population, and infer cellular developmental trajectories. To help users accomplish these goals, NUSeq assists users in every step of this process – from cell prep and sequencing library construction to bioinformatic analysis.
As the technology evolves, single-cell sequencing becomes more diverse to meet varying project needs. Currently, NUSeq offers multiple high-throughput single-cell sequencing platforms (10x Genomics, Illumina, and Parse Biosciences-now acquired by Qiagen), and one for low-throughput projects which need to sequence a small number of cells but at much greater sequencing depths. As of November 2025, single cell methylation sequencing is being established. Below is an overview of the established platforms at NUSeq:
- 10x Genomics Chromium iX
- Target cell number: Universal 3’ or 5’ gene expression—500-20,000 cells in each sample without multiplexing, or 500-5,000 cells in each sample with on-chip multiplexing; Flex gene expression (for fixed or archived samples)—up to 384 samples and 1 million cells can be multiplexed per channel (each chip has 8 channels); ATAC and Multiome (epigenomic profiling): 500-10,000 nuclei per sample
- Input type: fresh or frozen single-cell suspensions, fresh single-nucleus suspensions (including nuclei prepared from frozen tissues), fixed cells/nuclei and FFPE tissues (compatible with the Flex workflow)
- Applications: 3’ and 5’ single-cell (or nucleus) RNA-seq, T-cell and B-cell V(D)J clone profiling, cell surface and intracellular protein profiling, standalone single nucleus ATAC-seq and multiome (simultaneous single nucleus ATAC-seq and RNA-seq on the same cells)
- Illumina Single Cell Prep (previously known as PIP-seq)
- Target cell number: 100-100,000 cells in each sample
- Input type: high-quality single-cell (or nucleus) suspension prepared from mammalian fresh or cryopreserved cells obtained via cell culture, tissue dissociation, cell sorting, or other isolation methods. Alternatively, single-nucleus suspension prepared from frozen mammalian tissues is supported. Both fresh and fixed cells or nuclei are compatible
- Applications: 3’ single-cell (or nucleus) RNA-seq
- Parse Biosciences
- Target cell number: 10,000-1,000,000 cells, accommodating up to 384 samples
- Input type: fixed single-cell or nucleus suspension
- Application: single-cell (or nucleus) RNA-seq
- SMART-seq technology for manual low-throughput scRNA/scDNA-seq
- Target cell number: 1-100 cells
- Input type: 1-10 cells collected in individual tubes
- Usage: single-cell RNA-seq, single-cell DNA-seq
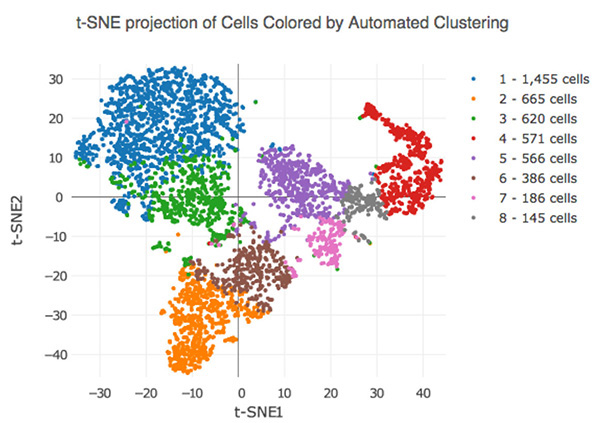
Unsupervised clustering of single-cell RNA-seq data processed using 10x Genomics Chromium. Human kidney cells are clustered based on similarities in gene expression, with each of the clusters representing a distinct subpopulation.
*Due to the high demand for scRNA-seq, it is important to schedule work well ahead of time to ensure processing.
The core’s 10x Genomics Chromium System is made available with an NIH S10 award. Please acknowledge Grant # 1S10OD025120 in publications.
Pricing
10x Genomics Chromium
- Single-cell (or nucleus) RNA-seq library preparation (cell/nucleus QC included) - $2,250 per sample (singleplex)
- Single-cell (or nucleus) RNA-seq library preparation with on-chip multiplexing - $3,200 per set of 4 samples
- Single nucleus ATAC-seq library preparation (cell/nucleus QC included) - $2,250 per sample
- Multiome (snRNA-seq + snATAC-seq) library preparation (cell/nucleus QC included) - $3,750 per sample
- Chromium Fixed cell single-cell (or nucleus) RNA-seq - Consult with NUSeq Core
Add-On Library Type
- Cell surface protein profiling add-on - $180 per sample
- V(D)J immune profiling add-on (applied to 5’ scRNA-seq only) - $450 per sample
Illumina Single Cell Prep
- $1,260 per sample, regardless of which kit (T2, T10, T20, or T100) is used. Kit costs are separate and billed as passthrough
Parse Biosciences
-
$1,400(WT Mini), $1750 (WT), or $2,100 (WT Mega) per kit for library preparation. Kit costs are separate and billed as passthrough
Manual Low-Throughput scRNA/scDNA-Seq
-
$420 per sample if <24 samples; $330 if >=24 samples
Sequencing
Cost of sequencing single-cell libraries varies with the platform chosen (see above), the total number of cells, the desired sequencing depth, etc. Cost projection for a particular project can be provided at the time of project consultation.
Please check the Core Pricing page for external rates.
Single-Cell Sequencing Service Details
Service Request
To initiate a single-cell sequencing project, please contact us. Project consultation is provided free-of-charge. Consultation with the core prior to starting a single-cell sequencing experiment is highly recommended to ensure accomplishment of project goals.
Single-cell sequencing services can be requested through NUcore.
Sample Submission
For all single-cell sequencing services, please make a sample submission appointment with us in advance.
- High-throughput single-cell sequencing platforms (10x Genomics, Illumina, and Parse): Users prepare fresh, frozen, or fixed cells (or nuclei) and submit them to NUSeq Core for sample QC and library construction.
- SMART-Seq platform: Users collect cell(s) in 0.2mL or 1.5mL tube in in 5µL of 1x PBS buffer and store at -80°C until sample submission to NUSeq Core.
Recommended Sequencing Parameters
10x Genomics Chromium Libraries
RNA-Seq Libraries: read length 28-10-10-90 bp — read 1 of 28 bp (cell barcode and UMI), i7 of 10 bp (sample index), i5 of 10 bp (sample index), and read 2 of 90 bp (transcript insert). Sequencing depth >20,000 reads per cell
ATAC-Seq Libraries: 50-8-16-50 bp — read 1 of 50 bp (transposed DNA), i7 of 8 bp (sample index), i5 of 16 bp (10x barcode) and read 2 of 50 bp (transposed DNA). Sequencing depth >25,000 reads per cell
Illumina Single Cell 3’ RNA Prep Libraries: 45-10-10-72 bp — read 1 of 45 bp (containing cell barcode), i7 of 10 bp (sample index), i5 of 10 bp (sample index), and read 2 of 72 bp (transcript insert, including intrinsic molecular identifiers or IMIs). Sequencing depth > 20,000 reads per cell
Parse Biosciences Libraries: 74-10-10-86 bp — read 1 of 74 bp (transcript insert), i7 of 10 bp (UDI), i5 of 10 bp (UDI), and read 2 of 86 bp (barcodes). Sequencing depth > 20,000 reads per cell
Manual Low-Throughput scRNA/scDNA-Seq: RNA-Seq — paired-end 50 bp reads at desired sequencing depth (e.g., 20-25 million reads per mammalian sample). DNA-Seq — Paired-end 150 bp reads often used at desired sequencing depth (e.g., 30x coverage for single-cell whole genome sequencing)
Bioinformatics
Data analysis is provided upon request.
Frequently Asked Questions
10x Genomics Chromium
What is the turnaround time?
From sample submission to library preparation it typically takes 2 weeks. It takes another 2-3 weeks for sample to be sequenced followed by up to 2 weeks for raw sequence demultiplexing and read alignment.
How many samples should I do at once?
For 10x Genomics platform, you can perform any number of samples. We usually recommend not to handle more than 4 samples at once.
What is the required cell (or nucleus) quality to proceed to single-cell/nucleus sequencing analysis?
For cell, we recommend the cell viability of >70%. For nucleus, please consult us to ensure the sample quality is good for 10x Genomics platform.
Can I integrate single-cell sequencing data with spatial transcriptomics data?
Yes. This is achievable. Please consult the core for more specifics.
Can I use long-read sequencer to sequence single-cell RNA-seq library?
Yes. This is achievable. Please consult the core for more specifics.
How do I start a single-cell sequencing experiment?
Please contact NUSeq (NUSeq@northwestern.edu) and we will help you through the whole workflow from sample preparation to library construction to data analysis.
Illumina Single Cell 3’ RNA Prep
How does the Illumina Single Cell 3’ RNA Prep work?
There are four steps in the workflow. First, cells are encapsulated into droplets along with barcoded hydrogel beads using an emulsification process, which is achieved with a vortexing process. Next, each cell is lysed in its droplet by heat-activated lysis reagents. This is followed by the capture of released mRNA molecules by bead-bound barcoded poly(T) oligonucleotides. The captured mRNA is then reverse transcribed into cDNA. The last step is preparation of sequencing libraries from the barcoded cDNA. The original method (called PIP-seq) is described in detail in Nature Biotechnology.
How many target cells can be analyzed?
There are four kit sizes available: T2, T10, T20, and T100, designed for profiling 2K, 10K, 20K, and 100K target cells per sample, respectively. Among these, the T2 and T10 kits are designed for smaller-scale projects. T2 is especially suited for pilot, organoid or stem cell studies, preliminary data collection for grant applications, and cell sorting optimization.
What human and mouse cell types have demonstrated success?
Human: Brain, Retina, Lung, PBMCs, Bone marrow, Adipocytes, Ovary, Testes, Umbilical cord, Lung organoid, Cerebral organoid, Epithelial cells, and mesenchymal cells.
Mouse: Brain, Retina, Inner ear hair cells, Heart, Mammary tissue, Liver, Intestine, PBMCs, Bone marrow, Plasma, Erythrocytes, Bladder, Embryonic tissue, Cerebral organoids, Xenografts, Brain tumor, and Neutrophils.
What other species have demonstrated success?
Pig (liver and spleen), Zebrafish, Killifish, Xenopus, Planarians, Skate, Tunicates, Sea anemone, Taxoplasma (a single-celled parasite), Yeast, Maize, and Bovine (ovary).
What is the multiplet rate?
Based on Illumina, the multiplet rate is less than 5%.
Can fixed tissues be used?
Yes.
Is there a detailed sample prep guide?
Please refer to the Illumina Single Cell Prep training packet, which breaks down the best practices for preparing your samples for the workflow.
Is there a cell size limit with Illumina Single Cell Prep?
Cells up to 60 µm in diameter have been successfully captured, although the exact upper limit has not yet been fully established. For cells ≥ 60 µm, Illumina recommends reducing cell input number to improve capture efficiency and minimize background noise. For cells > 80 µm, Illumina advises isolating nuclei and using them as input instead of whole cells.
How does this platform compare with other single cell sequencing platforms?
Each platform has its own unique characteristics in terms of cell recovery, sensitivity in detecting genes and transcripts, cell population identification, and differential gene expression profiling. Currently available benchmark data (e.g., Elz et al., 2025), while still limited, shows that PIP-seq is still behind the latest 10x Genomics chemistries in cell recovery and gene/transcript detection, but compares well with Parse.
Parse Bioscience
What is the turnaround time?
From sample submission to library preparation it typically takes 3 weeks. It takes another 2-3 weeks for sample to be sequenced followed by up to 2 weeks for raw sequence demultiplexing and read alignment.
How many samples should I do at once?
For Parse Bioscience platform, all fixed samples should be submitted to us all at once. For 100,000 cell and 1,000,000 cell kit, users can submit up to 48 and 96 samples, respectively.
What is the required cell (or nucleus) quality to proceed to single-cell/nucleus sequencing analysis?
For cell, we recommend using 1 million cells with cell viability of >70% prior to fixation. For nucleus, please consult us to ensure the sample quality is good.
Can I use long-read sequencer to sequence single-cell RNA-seq library?
Yes. This is achievable. Please consult the core for more specifics.
How do I start a single-cell sequencing experiment?
Please contact NUSeq (NUSeq@northwestern.edu) and we will help you through the whole workflow from sample preparation to library construction to data analysis.
Manual Low-Throughput scRNA/scDNA-Seq
What is the turnaround time?
What is the recommended cell collection method?
For sorted cell, please provide us 2-4 individual samples for protocol optimization. Please collect cell by sorter or manual pipetting in 0.2 mL tube with up to 3 µL of 1X PBS Buffer. Please store cells at -80°C until sample submission to NUSeq Core.
How do I start a single-cell sequencing experiment?
Please contact NUSeq (NUSeq@northwestern.edu) and we will help you through the whole workflow from sample preparation to library construction to data analysis.