Mitochondrial DNA Sequencing
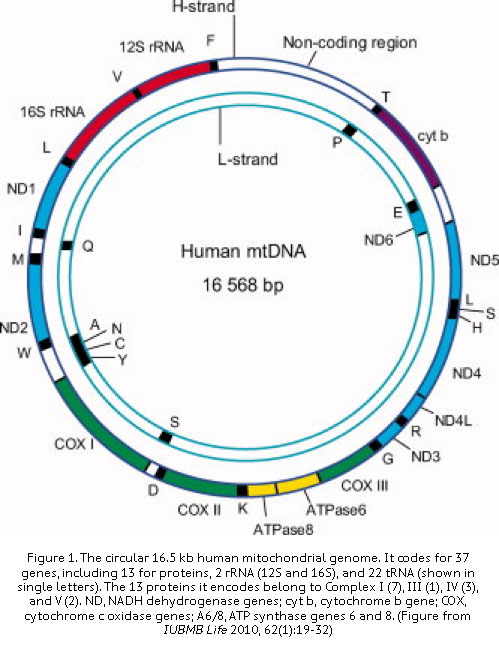
The mitochondrial genome is an independent genetic system in each eukaryotic cell outside the nuclear genome. While most researchers tend to overlook it because of its small size, the mitochondrial genome contains genes that are essential for cellular energetics and survival (Fig. 1). Because of the increased awareness on the importance of metabolism and bioenergetics in a wide variety of human diseases, more and more mitochondrial DNA (mtDNA) studies are performed to study the link between mtDNA sequence variation and disease development. Mitochondrial genome-wide association studies have established the connection between mtDNA and a wide variety of diseases such as cancer, autoimmune, and neurodegenerative disorders such as Alzheimer’s and Parkinson’s.
The close tie between mtDNA and cellular metabolic state also arises from the fact that mtDNA is highly subject to mutagenic attacks from reactive oxygen species released from the electron transport chain, leading to accumulation of somatic mutations. This, coupled with the very limited DNA damage repair capability in the organelle, leads to sequence alterations in mtDNA at a much higher frequency than that in the nuclear DNA. As each mitochondrion contains 2-10 copies of mtDNA and each cell contains hundreds to thousands copies, the presence of heterogeneous mtDNA sequences, due to different mutations in different copies, results in a condition called heteroplasmy. Increased levels of mtDNA heteroplasmy, as an indicator of cellular metabolic dysfunction, in turn leads to elevated metabolic abnormality due to amino acid coding change in key oxidative phosphorylation proteins. This vicious cycle is the basis of an increasing list of human diseases.
NUSeq provides mtDNA-seq for detection of specific mtDNA mutations associated with a disease, determination of mtDNA heteroplasmy, and haplogroup classification (Fig. 2). The process starts from extracted genomic DNA, which contains both mtDNA and nuclear DNA, without the need to purify mitochondria. Enrichment of mtDNA is achieved through selective amplification of the mitochondrial genome with long-range PCR. The enriched mtDNA is then subjected to library prep and then sequencing.
mtDNA-seq Library Prep Pricing
Library construction cost for mtDNA-seq is listed on the Core Pricing page.
Mitochondrial DNA Sequencing
Service Request
Project consultation is provided free-of-charge. RNA-seq services can be requested through NUcore. Please finish and submit our project intake form when requesting service.
Sample Submission
Total genomic DNA is the starting material for mtDNA-seq. There is no need to isolate mitochondria for mtDNA extraction. Virtually all DNA extractions contain both nuclear DNA and mtDNA. The quality of DNA affects the enrichment of mtDNA by long-range PCR. Highly pure, freshly prepared, and high molecular weight gDNA is preferred. The required amount is at least 10 ul gDNA in 10 ng/ul concentration.
After sample dropoff, core staff conducts sample QC, which includes Qubit concentration measurement and Bioanalyzer fragment analysis, prior to mtDNA enrichment and library construction.
Sequencing Mode
Single-end 50 or 75bp reads are enough for mapping mtDNA reads to the mitochondrial reference genome. Because of the small size of the genome, usually there is more than enough coverage even with heavy multiplexing.
Bioinformatics
Data analysis is provided upon request. Standard mtDNA-seq bioinformatics service includes sequencing data QC, alignment to the reference mitochondrial genome, and variant analysis. Heteroplasmy and/or haplogroup analyses are available for request. Raw data and analysis results are usually returned to user via OneDrive.
Bioinformatics is billed on a per hour basis. Usually the time spent on a typical project for standard analysis is averaged at one hour per sample.
Frequently Asked Questions
Do I need specific DNA extraction kit or procedure to purify genomic DNA that contains mtDNA?
Nearly all DNA extraction kits or procedures purify both nuclear and mitochondrial DNA into the final product. No particular kit is needed.
Does the long-range PCR performed to enrich mtDNA from total gDNA amplify NUMTs (nuclear mitochondrial DNA sequences)?
NUMTs are a result of insertion of mtDNA sequence into the nuclear genome. While there are numerous NUMTs existing in the nuclear DNA, the primers we use for mtDNA enrichment is highly specific to avoid any potential amplification of NUMTs from nuclear DNA. The primers are in silico and experimentally validated, and used by the mtDNA research community.
Can I use NGS data from other projects, such as exome-seq, RNA-seq, ChIP-seq, or ATAC-seq, for mtDNA sequence analysis?
Besides each cell contains usually only two sets of nuclear DNA but hundreds to thousands copies of mtDNA, mtDNA reads are usually present in sequencing data that are not intended for mtDNA analysis, including the NGS applications list above. Sometimes the proportion of mtDNA reads is substantial. Therefore, it is possible to repurpose sequencing data generated from these applications for mtDNA analysis.