Technologies
Breakthroughs in modern biomedical research are largely driven by rapidly evolving genomics technologies. The NUSeq Core is a state-of-the-art core facility that provides genomics and bioinformatics support to investigators at Northwestern University, affiliated institutions, and external academic and commercial institutions. The major technologies available at the core are listed below:
Short Read Sequencing
Applications: Whole Genome/Exome Sequencing, Gene Panel Sequencing, RNA-Seq (incl. Single Cell RNA-Seq), ChIP-Seq, Methyl-Seq, Microbiome Sequencing
With the power to sequence millions to hundreds of millions of DNA fragments simultaneously in a matter of hours or few days, NGS has been the major driving force in life science and medicine in the last decade. The sequences (or reads) generated from the DNA molecules can be used to assemble new genomes, while more often they are used for mutation or variation detection, differential gene expression analysis, detection of protein-DNA interaction, epigenomics, and environmental microbial diversity profiling.
Northwestern investigators continually pioneer new strategies to solve their research questions with NGS. To meet the sequencing needs of our users, NUSeq maintains a fleet of major NGS systems, summarized below. If your project requires a different NGS platform, please notify us as we are constantly working to build our capacity.

Illumina NovaSeq X Plus
- Top data throughput, cost effective
- Three flow cell types at different throughput levels:
- 25B flow cell: 2,000-2,500 million* reads per lane, 8 lanes per flow cell. Read length available: 2x150 bases. Sequencing on this flow cell is the most cost effective among all NUSeq sequencers at <$3 per Gbp
- 10B: 900-1,000 million reads per lane, 8 lanes per flow cell. Read length available: 2x50 and 2x150 bases
- 1.5B: 600-700 million reads per lane, 2 lanes per flow cell. Read length available: 2x50 and 2x150 bases
- Visit the Illumina NovaSeq X Plus Specifications page for more technical specifications.

Element AVITI
- Intermediate throughput, quick turnaround time
- Read length and output:
- 2x75 bases: 400-500 million reads in Medium Output mode, or 800-1,000 million reads High Output, per flow cell. Each flow cell has 2 lanes
- 2x150 bases: 400-500 million reads in Medium Output mode, or 800-1,000 million reads High Output, per flow cell. Each flow cell has 2 lanes
- 2x300 bases: 100 million reads in Medium Output mode, or 300 million reads High Output, per flow cell. Each flow cell has 2 lanes
- Accuracy: Equivalent to other short read sequencing platforms. UltraQ mode achieves Q50 accuracy in 70% of reads, and Q40 in 90% of reads, for applications that require highest accuracy possible
- Visit the Element Biosciences AVITI page for more technical specifications.

Complete Genomics DNBSEQ-G400
- Low to intermediate throughput, quick turnaround time
- Two flow cell types for flexibility
- FCS (small): 200-250 million reads per lane, 2 lanes per flow cell
- FCL (large): 400 million reads per lane, 4 lanes per flow cell
- Read length available: FCS, 2x150 bases; FCL, 2x100 and 2x150 bases. The specific read length required by STOmics Stereo-seq is also available.
- Visit the Complete Genomics DNBSEQ-G400 page for more technical specifications.

Illumina NextSeq 500
- Intermediate throughput, quick turnaround time
- 130 or 400 million reads from one flow cell depending on sequencing mode, no separable lanes
- Read length available: 1x75, 1x150, and 2x75 bases
- Visit the Illumina NextSeq 550 page for more technical specifications.

Illumina MiSeq
- Low throughput, quick turnaround time
- 1, 4, 15, to 25 million reads from one flow cell with throughput depending on flow cell type, no separable lanes
- Read length available: 2x150, 2x250, and 2x300 bases
- Visit the Illumina MiSeq page for more technical specifications.
Long Read Sequencing
Applications: Full-Length Transcript Profiling (at both bulk and single cell levels), Splicing Isoform Detection, Fusion Transcript Detection, Whole Genome Assembly, Haplotype Resolution, Structural Variant Detection, DNA Repeat Expansion Determination, Native Methylation Calling, Metagenomics
Long read sequencing technologies overcome challenges and limitations of short-read sequencing, and enable investigators to resolve complex genomic regions, accurately assemble genomes, identify structural variants, and unravele gene alternative splicing isoforms. Below are two currently available long read sequencing platforms available at NUSeq, both of which have seen significant advancements in recent years and continue to improve on accuracy and throughput.
Oxford Nanopore Long Read Sequencer
- Read length: 10-100 kb for long-read sequencing, and 100-300 kb for ultra-long read sequencing mode. 4 Mbp is the current record
- Read accuracy: Q20-Q30
- Data throughput:
- PromethION: We see typically 50-100 Gb data per flow cell
- MinION: We see typically 10-20 Gb data per flow cell
- Simultaneous detecton of epigenetic modifications with primary sequence
- Visit the Oxford Nanopore Technologies PromethION and MinION page for more technical specifications.

PacBio Revio HiFi Long Read Sequencer
- Read length: Typically in the 15-20 kb range
- Read accuracy: Q30
- Data throughput: 90 Gb data per SMRT cell
- Generates methylation (5mC at CpG sites) data from native DNA simultaneously
- Visit the PacBio Revio page for more technical specifications.
Single-Cell Sequencing
Applications: Single-Cell RNA-Seq, Single Cell ATAC-Seq, Single-Cell DNA-Seq, Single-Cell Methyl-Seq, Single-Cell Multi-Omics
Single cell sequencing offers unprecedented opportunities to study cell-to-cell variation, identify/visualize different cell types/identities in a population, and infer cellular developmental trajectories. NUSeq offers multiple single-cell sequencing platforms, including 10x Genomics and Parse Biosciences, to meet the increasing needs for single cell sequencing, As listed above, besides transcriptome, single cell sequencing increasingly encompasses genome, exome, and epigenome of individual cells. While most single cell sequencing projects analyze large numbers of cells, the Core also offers a low-throughput option for teams who need to sequence small numbers of cells at greater depth.

10x Genomics Chromium
- Target cell number: up to 20,000 cells/nuclei in each sample
- Input type: Freshly prepared single-cell (or nucleus) suspension, fixed cells (or nuclei), cryopreserved cells, and FFPE embedded tissues
- Available assays: 3’ gene expression, 5’ gene expression, immune repertoire profiling, chromatin accessibility, multiome (i.e., simultaneous chromatin accessibility and gene expression from the same set of nuclei)
- Visit our Single-Cell Sequencing page for service details.
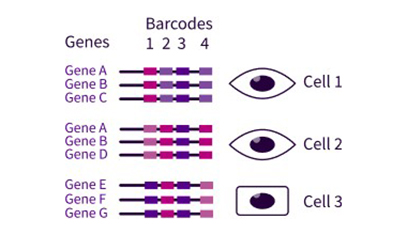
Parse Evercode
- Target cell number: 100,000-1,000,000 cells in each sample
- Input type: Fixed single-cell or -nucleus suspension
- Available assays: whole transcriptome, TCR profiling, CRISPR detect, gene capture
- Visit our Single-Cell Sequencing page for service details.
Other single cell platforms are also available, e.g., those from Bioskryb Genomics for integrated genome/transcriptome analysis and Scale Biosciences for single cell methylation studies.
Spatial Genomics
Applications: Spatially Resolved Transcriptomics, Tissue Neighborhood Analysis, Tumor Microenvironment Analysis, Cell-Cell Interaction Inference, Spatial Cell Atlasing
Spatial context is important for cells to carry out their functions. For diseased conditions, pathologists examine tissue sections to look for microenvironments where cells turn abnormal. Spatial genomics provides a “map” of gene activity in morphology-preserved tissues, often with subcellular resolution. This state-of-the-art technology enables us to investigate gene expression in the context of tissue structure to shed light on functional diversity and heterogeneity in regions of interest. Depending on technical approaches employed, spatial resolution of gene transcripts in tissue sections is achieved using a sequencing-based workflow or in situ hybridization.

Visium HD (10x Genomics)
- Next-gen sequencing based, whole transcriptome
- Resolution: 2 µm
- Tissue capture area size: 6.5 x 6.5 mm (each Visium slide has two capture areas)
- Sample type: FFPE and fresh frozen tissues
- Enables integration of spatial gene expression data with bright-field and fluorescence microscope images
- Visit our Spatial Transcriptomics page for service details.

Xenium (10x Genomics)
- In situ hybridization based, targeted analysis
- Resolution: 200 nm
- Tissue image area size: 10.45 x 22.45 mm
- Sample type: FFPE and fresh frozen tissues
- Allows immunofluorescence, H&E staining, or Visium whole transcriptome analysis on the same tissue section
- Visit our Spatial Transcriptomics page for service details.
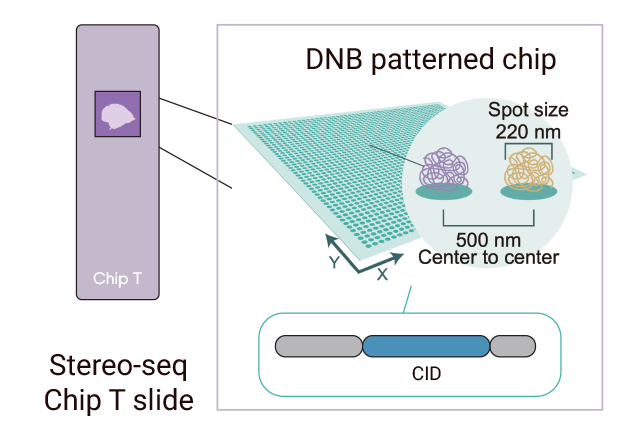
Stereo-Seq (STOmics)
- Sequencing based, whole transcriptome analysis
- Spot size and pitch: Each transcript capture spot is 0.22 µm in diameter, with spot center-to-center distance of 0.5 µm
- Tissue capture area size: ranging from 5 mm x 5 mm to custom-designed chips as large as 13 cm x 13 cm
- Sample type: FFPE and fresh frozen tissues
- Visit our Spatial Transcriptomics page for service details.
Microarray
Applications: Genome-Wide Genotyping, Epigenome-Wide DNA Methylation Profiling
Microarray was the first high-throughput genome technology developed based on hybridization of pre-designed nucleic acid probes to molecules present in user samples. Signal readouts from the hybridization are then used to analyze gene expression, gene sequence variation, DNA methylation, etc. Currently most NUSeq users employ this time-tested technology for genome-wide genotyping and methylation analyses. Illumina genotyping arrays can be used in genome-wide association studies (GWAS) to establish relationships between nucleotide/structural changes and development of diseases (or biological traits) in a variety of species. For epigenome-wide association studies (EWAS), the Illumina human methylationEPIC and mouse methylation arrays are used to identify differentially methylated cytosines at single nucleotide resolution across the genome.

Illumina Genotyping Arrays
- Available arrays:
- Human Global Diversity Array (GDA) (shown above) – Built on a multi-ethnic backbone, optimized for crosspopulation imputation coverage of the genome, and enables polygenic score development. Used by the All of Us Research Program
- Human Global Screening Array (GSA) – A cost effective array for population genetic studies, variant screening, and precision medicine research. Contains optimized multiethnic and high-value disease-associated variants
- Visit the Illumina Whole-Genome Genotyping page for more genotyping arrays for human and other species.
- Array contents:
- GDA – over 1.8 million markers across the genome, encompassing variants with known disease associations and pharmacogenomics markers. Can add up to 175K custom markers;
- GSA – 654K fixed markers. With capacity to add up to 100K custom markers;
- Contents for other genotyping arrays are available on the Illumina genotyping array site above.
- Number of samples per chip: Varies with array type; 8 for GDA and 24 for GSA
- Sample input: 200 ng DNA (including those extracted from FFPE tissues)

Illumina Methylation Arrays
- Available arrays: Human MethylationEPIC v2.0 BeadChip (shown above, backwards compatible with HumanMethylation450 and MethylationEPIC v1.0 chips), Mouse Methylation BeadChip
- Array contents: The Human MethylationEPIC v2.0 array covers over 935,000 annotated methylation sites across the genome. The Mouse Methylation array covers over 285,000 sites. The covered sites on these arrays include those located in CpG islands, non-CpG areas, transcription start sites, enhancers and super-enhancers, imprinted loci, gene body regions, differentially methylated regions in cancer, differentially accessible chromatin regions in cancer, etc
- Number of samples per chip: 8 (Human MethylationEPIC v2.0 array), 12 (Mouse Methylation array)
NanoString nCounter
Applications: Targeted gene expression profiling
The NanoString nCounter Sprint Profiler is a platform for multiplex analysis of up to 800 RNA or DNA targets. This platform is based on molecular hybridization using target-specific probe pairs, and the digital readout of each target is achieved by NanoString’s unique fluorescence barcoding and single molecule imaging. This system is designed for studies that interrogate gene expression in a particular pathway or biological process. In addition to the catalogued gene panels (or CodeSets) below, users have the option of designing custom panels based on specific needs. The system also enables detection of gene fusions using junction probes, analysis of miRNAs, and simultaneous measurement of DNA/RNA/proteins.
- Predesigned panels: Cardiovascular Disease, Cell and Gene Therapy, Immunology, Infectious Disease, Neuroscience, and Oncology
- Number of samples processed per cartridge: 12
- Input required: 50 ng total RNA if extracted from fresh cells or flash frozen tissue; 150 ng if from FFPE tissue. Low RNA input (needs amplification prior to hybridization): 1 ng if extracted from fresh cells or flash frozen samples; 10 ng RNA if from FFPE tissue.
Please visit the NanoString service page for more details.
Digital and Quantitative PCR
Applications: Detection of DNA Target (e.g., ctDNA or pathogenic DNA), CNV, Target Gene Expression, CRISPR Genome Editing Screening, NGS Library QC, NGS Discovery Validation
Droplet Digital PCR (ddPCR)
Digital PCR is the latest generation of PCR technology for absolute quantification of target DNA/RNA molecule(s), with significantly improved sensitivity and accuracy. This technology is especially suitable for targeted detection of circulating tumor DNA in liquid biopsy, rare somatic DNA mutations, or pathogenic nucleic acids at levels below the current detection limit. The detection sensitivity can reach 1 mutation in 1 million human genomic DNA copies, or 1 bacterial/viral DNA copy in 500,000 human cells.
NUSeq maintains the Bio-Rad QX200 ddPCR System, which has the capability to generate 20,000 individual PCR reaction droplets from a 20 ul PCR reaction. After PCR, all droplets are examined by the droplet reader for presence or absence of amplification signal. The percentage of positive droplets are then used to calculate the absolute copy number of the target DNA/RNA molecule(s) in the original sample on the basis of Poisson statistics.

(above) The Bio-Rad QX200 ddPCR System and the steps of carrying out a ddPCR experiment.
Quantitative Real-Time PCR (qPCR)
NUSeq maintains a QuantStudio 7 Flex system (ThermoFisher) to meet users’ project needs for real-time PCR technology. This system provides interchangeable block formats (96-well fast, 384-well, and TaqMan array card block) and sufficient multiplexing capabilities. This system is equipped with streamlined software, responsive touch-screen, automation capabilities, and easy block change without the need for any tools.

Cell Line Authentication
Applications: Cataloged Cell Line Confirmation, Patient-Derived Cell Line Matching
Verifying the identity of a cell line used in a project is essential to maintain scientific rigor and experimental reproducibility. NUSeq achieves authentication of human cell lines through an STR (Short Tandem Repeats) profiling assay. By analyzing a relatively small number of STR loci, this assay provides enough power to discriminate different human cell lines.
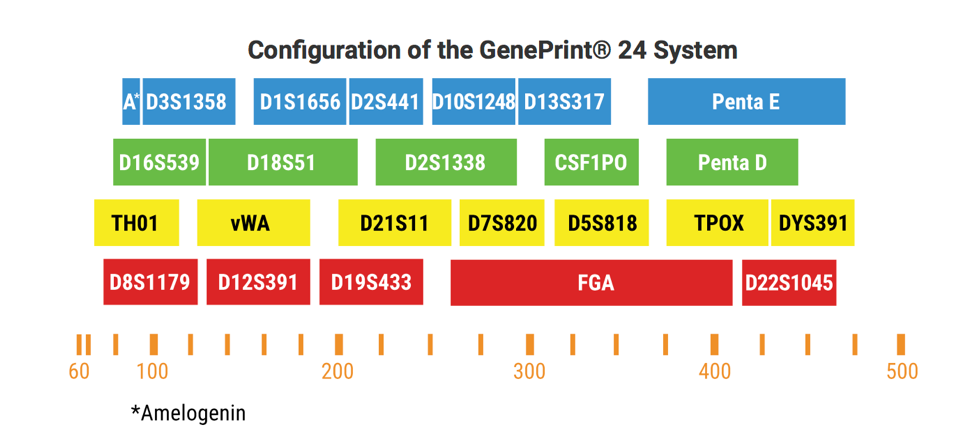
NUSeq uses the GenePrint 24 System from Promega for our STR-based cell line authentication. This system is based on co-amplification and five-color detection of 24 STR loci as shown below.
DNA/RNA Sample Prep and QC
Applications: DNA and RNA Extraction (prior to downstream analyses including sequencing, array processing, PCR, etc.)
Automated or Manual DNA/RNA Extraction
High-throughput DNA/RNA extraction is offered on automated platforms (see below) to accommodate projects that submit large numbers (e.g., thousands) of samples. For samples that need special attention, manual extraction is also provided. NUSeq has experience extracting DNA, RNA, or both (i.e., co-purification), from a variety of sources, including FFPE sections, flash frozen tissues, whole blood, buffy coat, serum/plasma, saliva, buccal swabs, and cultured cells.

Chemagic 360 from PerkinElmer (now Revvity)
- Based on chemagen’s M-PVA Magnetic Bead technology
- Ideal for larger volume samples like saliva or whole blood
- Input volume: up to 4 ml if liquid (blood, saliva, etc.)
Maxwell RSC 48 from Promega
- Uses Promega’s cartridge-based Paramagnetic Particle Moving technology
- Ideal for high-throughput projects with smaller sample sizes
- Input volume: up to 400 µl if liquid (blood, saliva, etc.)
Manual extraction is also available upon request. For more details on the Core’s DNA/RNA services, please visit the DNA/RNA Extraction Service page.
DNA/RNA Sample QC
Core-prepared or user-submitted samples are qualified and quantified using Bioanalyzer, TapeStation, Fragment Analyzer, and Qubit. Sequencing libraries are also quantified using qPCR. Please visit the DNA/RNA Quality Control page for more information.