NanoString for Highly Multiplexed Target Gene Analysis
The NanoString nCounter Sprint Profiler — a new addition to NUSeq (as of September 2020) — is a platform for multiplex analysis of up to 800 RNA or DNA targets. For common pathways or bio-functions, NanoString provides a number of ready-to-use, predesigned gene panels, including those for oncology, immunology, and neurobiology. In addition to the catalogued gene panels, NUSeq Core users have the option of designing custom panels based on specific needs. This system is ideal for studies that interrogate genes in a particular pathway or bio-function, or to validate expression pattern of a group of genes after global gene expression profiling with RNA-seq. The system also enables detection of gene fusions using junction probes, analysis of miRNAs, and simultaneous measurement of DNA/RNA/proteins.
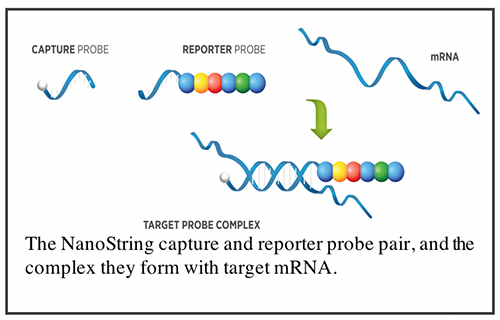
The NanoString System achieves digital read-out of up to 800 targets based on the use of target-specific hybridization probe pairs (see the figure below). Each probe pair contains a reporter probe that carries a unique fluorescence barcode, and a capture probe that is biotinylated for immobilization (or capture) of the target-probe complex to a streptavidin-coated slide for scanning and signal collection. Both probes contain sequences that are specific to its designated target. There is no enzymatic step in this process, reducing the chance of introducing bias. Through the unique fluorescence barcoding scheme and single molecule imaging, gene targets are quantified in a single reaction at high sensitivity. Its super sensitivity means limited sample input, such as patient biopsy, is feasible, and single cell profiling at the pathway level is achievable with minimal amplification.
NanoString Pricing
Sample hybridization from submitted input sample (extracted RNA or cell lysate) is $150 per cartridge (12 samples). This step can be performed in user labs to keep cost down.
Data generation on the SPRINT Profiler from hybridized samples is $480 per cartridge (12 samples).
NanoString gene panels, either predesigned or custom, are not included and need to be ordered separately. Users can order them from NanoString directly, or NUSeq can purchase them and bill as a passthrough.
NanoString
Service Request
Project consultation is provided free-of-charge. NanoString services can be requested through NUcore.
Sample Submission
The recommended amount of total RNA for CodeSet hybridization is:
- If extracted from fresh cells or flash frozen tissue: 50 ng (~10 ng/μl)
- If extracted from FFPE tissue: 150 ng (~30 ng/μl)
Bioinformatics
The quantification of each target gene is quite straightforward and achieved through counting the total number of unique fluorescence barcodes attached during sample hybridization. As a result, the data from the Profiler is human readable. To facilitate data analysis, NanoString offers the free nSolver software. Additional open-source packages can also be used for advanced analyses. If needed, NUSeq’s bioinformatics group can provide help on data analysis.
Frequently Asked Questions
Do I need to run technical replicates?
Running technical replicates is usually not necessary. Biological replicates should be run whenever possible.
What endogenous controls are included in each run?
Fourteen ERCC controls are included in each CodeSet. Six of them are positive hybridization controls, and eight are negative controls.
How should I determine the quality and quantity of my RNA?
RNA quality should be determined using a Bioanalyzer or TapeStation. A fluorometer, such as Qubit, should be used to quantify RNA for better accuracy.
Would DNA contaminants interfere with the results?
DNA molecules are expected to remain double-stranded during sample hybridization conditions. As a result, they should be invisible to the probes and should not interfere with RNA hybridization to the probes.
What if I have less input material than the recommended amount?
Good results can be obtained with less material from 1 or 10 ng RNA extracted from fresh frozen or FFPE samples, respectively. Such low amount of input RNA needs to be amplified using the nCounter Low RNA Input Amplification Kit prior to hybridization.